Chemical Biology
Functional Metabolomics and Lipidomics
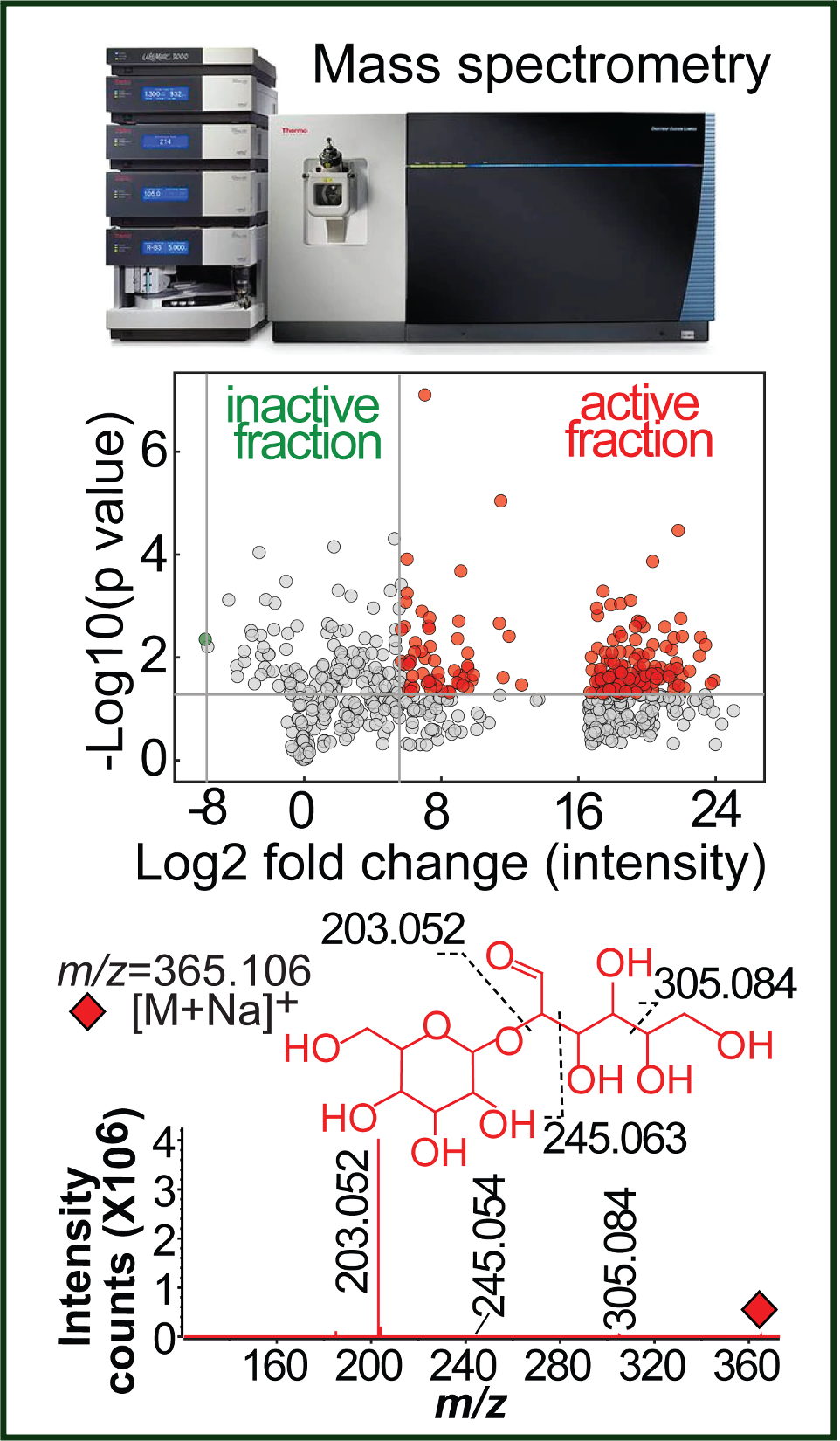 We are establishing metabolomic and lipidomic platforms to profile and identify various metabolites and lipids that bind and interact with antigen-presentation molecules and other proteins. Metabolites can be defined as polar or non-polar small molecules with mass units generally less than 2000 and as direct products of various metabolic pathways (human metabolome database). More narrowly, water-soluble metabolites are sometimes designated “metabolites” to separate them from lipids. Small lipids and metabolites are critical compounds derived from various metabolic pathways to control and regulate immune pathways through binding and interacting with antigen-presentation molecules, transcription factors, hormone receptors, and other proteins. We will set up the metabolomic capability to profile and identify biologically important compounds in immune responses, microbial infections, and cancers. We will also integrate bioassay-guided isolation, serial or 2D high-performance liquid chromatography, and other chemical and biochemistry techniques for compound preparation and identification.
We are establishing metabolomic and lipidomic platforms to profile and identify various metabolites and lipids that bind and interact with antigen-presentation molecules and other proteins. Metabolites can be defined as polar or non-polar small molecules with mass units generally less than 2000 and as direct products of various metabolic pathways (human metabolome database). More narrowly, water-soluble metabolites are sometimes designated “metabolites” to separate them from lipids. Small lipids and metabolites are critical compounds derived from various metabolic pathways to control and regulate immune pathways through binding and interacting with antigen-presentation molecules, transcription factors, hormone receptors, and other proteins. We will set up the metabolomic capability to profile and identify biologically important compounds in immune responses, microbial infections, and cancers. We will also integrate bioassay-guided isolation, serial or 2D high-performance liquid chromatography, and other chemical and biochemistry techniques for compound preparation and identification.
Representative Publications
Metabolites and Lipids as T cell Antigens and Protein Ligands
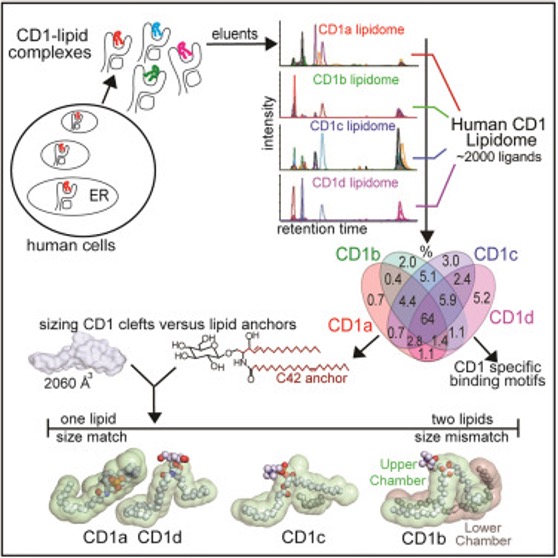 Antigen-presenting molecules bind peptide, lipid, or metabolite ligands to interact with T cell receptor (TCR) and activate different T cell populations. Antigen-presenting molecules, including MR1 and CD1, bind metabolite and lipid ligands to interact with MAIT cells and CD1-restricted T cells, respectively. To identify the metabolites and lipids for T cell activation, we expressed and isolated non-peptidic ligands from MR1 and CD1 proteins to characterize their structures and functions for T cell activation. Taking the CD1 system as an example, human CD1a, CD1b, CD1c, and CD1d proteins bind lipid antigens for surface display to T cells. We solved lipidomes interacting with these four human CD1 proteins and provided a map of self-lipid display via detecting >2,000 CD1-lipid complexes and demonstrating a broad presentation of self-sphingolipids and phospholipids. These data allowed further size-matching of lipids with CD1 lipid-binding clefts and provided candidate lipid blockers and agonists for T cells.
Antigen-presenting molecules bind peptide, lipid, or metabolite ligands to interact with T cell receptor (TCR) and activate different T cell populations. Antigen-presenting molecules, including MR1 and CD1, bind metabolite and lipid ligands to interact with MAIT cells and CD1-restricted T cells, respectively. To identify the metabolites and lipids for T cell activation, we expressed and isolated non-peptidic ligands from MR1 and CD1 proteins to characterize their structures and functions for T cell activation. Taking the CD1 system as an example, human CD1a, CD1b, CD1c, and CD1d proteins bind lipid antigens for surface display to T cells. We solved lipidomes interacting with these four human CD1 proteins and provided a map of self-lipid display via detecting >2,000 CD1-lipid complexes and demonstrating a broad presentation of self-sphingolipids and phospholipids. These data allowed further size-matching of lipids with CD1 lipid-binding clefts and provided candidate lipid blockers and agonists for T cells.
